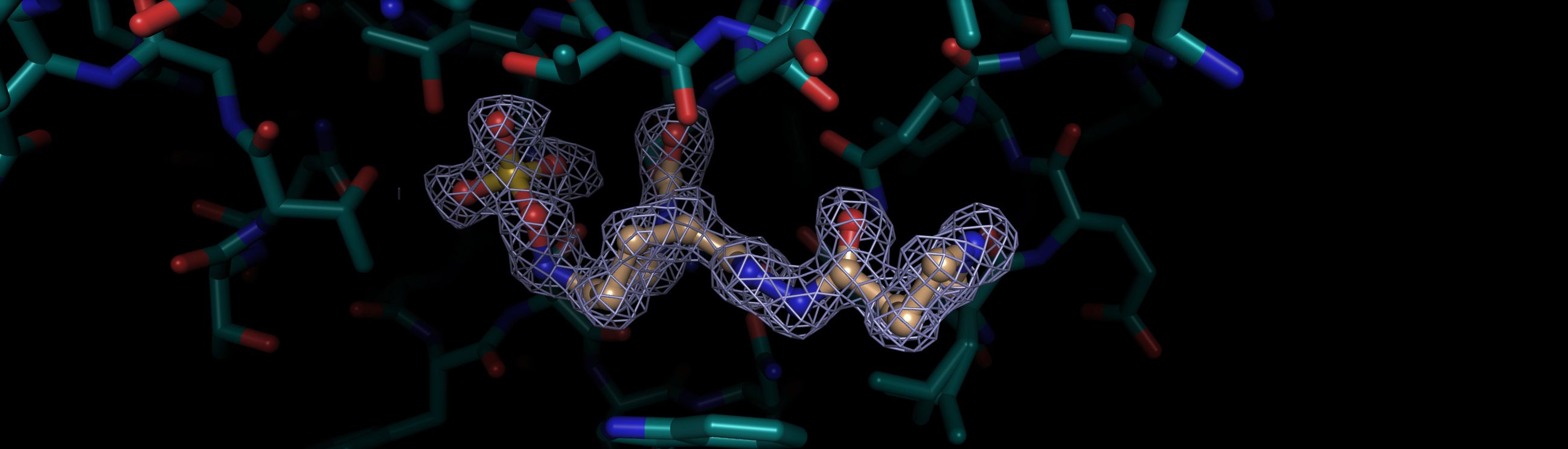
Structural Biology And Drug Discovery Approaches On Bacterial Cell Wall Targets

Structural Biology And Drug Discovery Approaches On Bacterial Cell Wall Targets
2/16/21
https://pubmed.ncbi.nlm.nih.gov/33593978/
This work was in collaboration with Wockhard Research Center and Dr. Robert Bonomo.
6/20/20
https://pubmed.ncbi.nlm.nih.gov/32420736/
This work was in collaboration with Drs. Robert Bonomo and Mark Plummer.
7/1/20
3/9/20
https://www.ncbi.nlm.nih.gov/pubmed/32152075
This work was in collaboration with Basilea Pharmaceutica International Ltd. and Dr. Robert Bonomo.
12/30/19
12/1/19
https://www.osc.edu/sites/default/files/page-files/2019%20Research%20Report.pdf
This work featured an all-atom 1-microsecond molecular dynamics simulation
of the doughnut-shaped lytic transglycosylase enzyme and its peptidoglycan substrate and was
published last year.
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0197136
11/8/19
https://aac.asm.org/content/early/2019/11/05/AAC.01841-19.long